Remember you should
add code chunks by clicking the Insert Chunk button on the toolbar or by pressing Ctrl+Alt+I to answer the questions!
render your file to produce a markdown version that you can see!save your work often
commit it via git!push updates to github
Examples
If interaction is significant
Following the memory example from class, read in and check data
<- read.table ("http://www.statsci.org/data/general/eysenck.txt" , header = T,stringsAsFactors = T)str (memory)
'data.frame': 100 obs. of 3 variables:
$ Age : Factor w/ 2 levels "Older","Younger": 2 2 2 2 2 2 2 2 2 2 ...
$ Process: Factor w/ 5 levels "Adjective","Counting",..: 2 2 2 2 2 2 2 2 2 2 ...
$ Words : num 8 6 4 6 7 6 5 7 9 7 ...
Let’s put younger level first
library (plyr)$ Age <- relevel (memory$ Age, "Younger" )
and graph
Loading required package: lattice
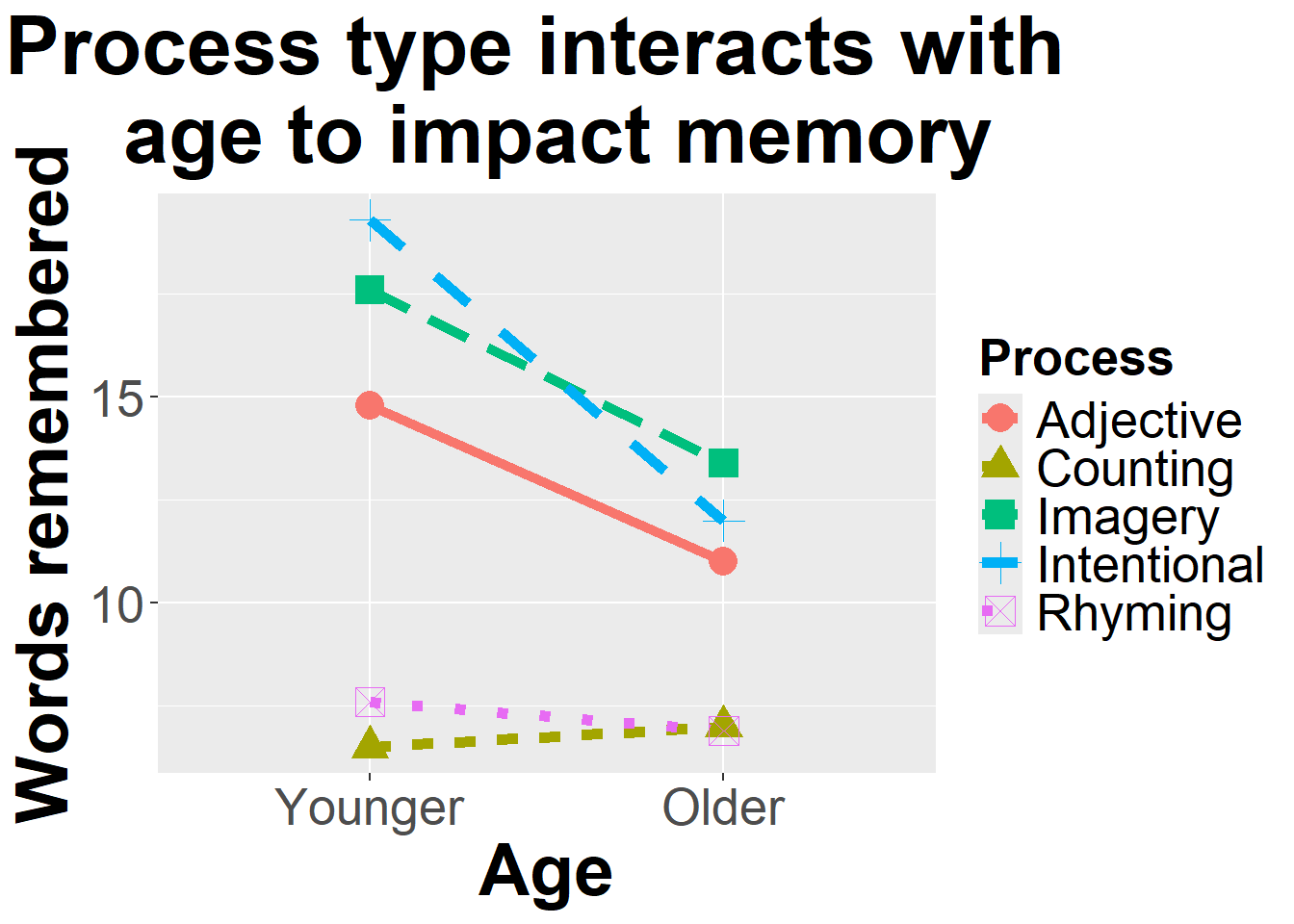
<- summarySE (memory, measurevar= "Words" , groupvars = c ("Age" , "Process" ), na.rm = T)library (ggplot2)ggplot (function_output, aes (x= Age, y= Words,color= Process, shape = Process)) + geom_line (aes (group= Process, linetype = Process), size= 2 ) + geom_point (size = 5 ) + ylab ("Words remembered" )+ xlab ("Age" ) + ggtitle ("Process type interacts with \n age to impact memory" )+ theme (axis.title.x = element_text (face= "bold" , size= 28 ), axis.title.y = element_text (face= "bold" , size= 28 ), axis.text.y = element_text (size= 20 ),axis.text.x = element_text (size= 20 ), legend.text = element_text (size= 20 ),legend.title = element_text (size= 20 , face= "bold" ),plot.title = element_text (hjust = 0.5 , face= "bold" , size= 32 ))
Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
There appears to be some interactions. Let’ build a model
<- lm (Words ~ Age * Process, memory)
and check assumptions.
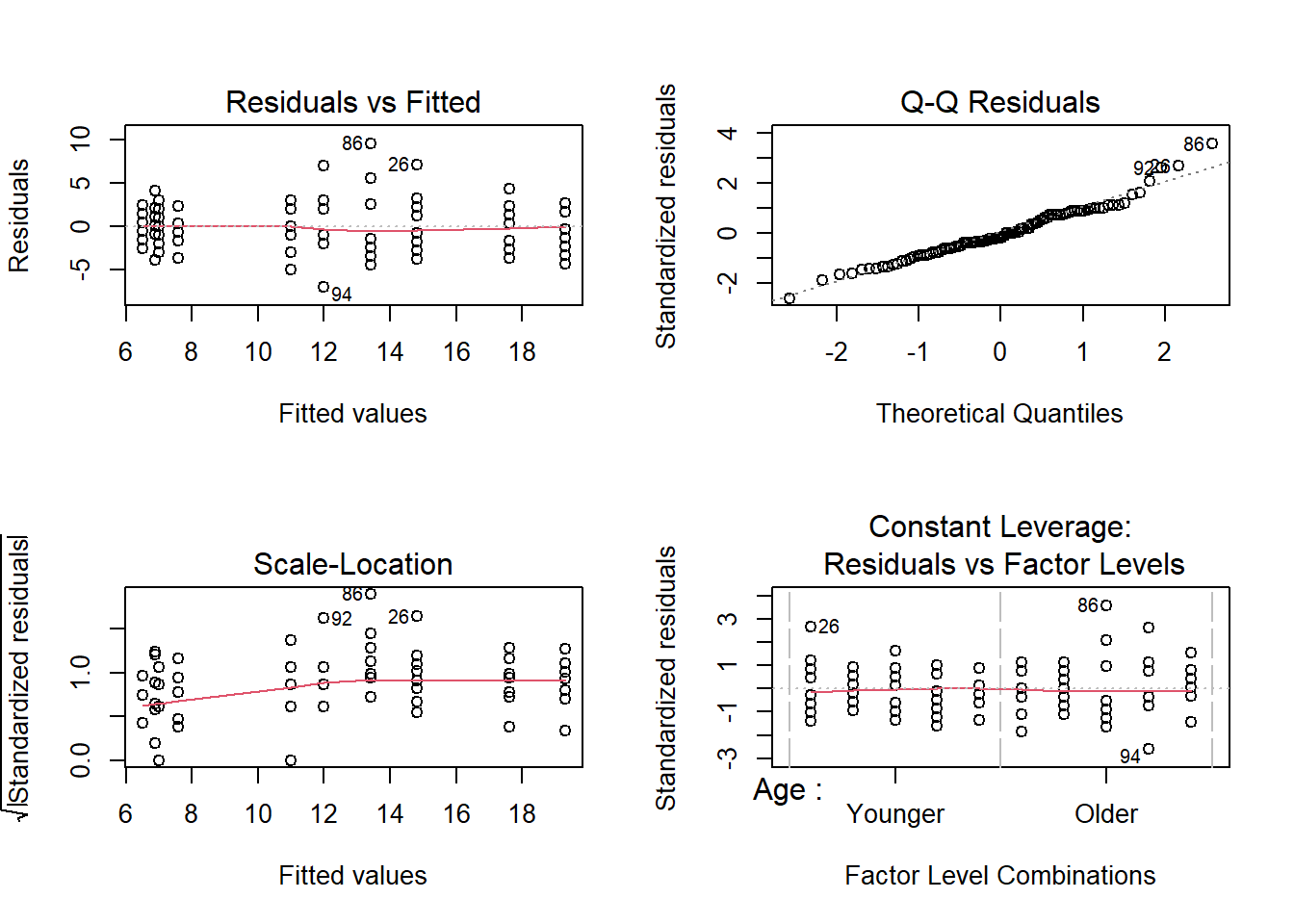
par (mfrow= c (2 ,2 ))plot (memory_interactions)
These appear to be met, so look at output
Warning: package 'car' was built under R version 4.4.1
Loading required package: carData
Anova (memory_interactions, type = "III" )
Anova Table (Type III tests)
Response: Words
Sum Sq Df F value Pr(>F)
(Intercept) 2190.4 1 272.9281 < 2.2e-16 ***
Age 72.2 1 8.9963 0.0034984 **
Process 1353.7 4 42.1690 < 2.2e-16 ***
Age:Process 190.3 4 5.9279 0.0002793 ***
Residuals 722.3 90
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Since interaction is significant, analyze subsets. For example,
<- lm (Words ~ Process, memory[memory$ Age == "Younger" ,])plot (memory_interactions_young)Anova (memory_interactions_young, type = "III" )
Anova Table (Type III tests)
Response: Words
Sum Sq Df F value Pr(>F)
(Intercept) 2190.4 1 343.442 < 2.2e-16 ***
Process 1353.7 4 53.064 < 2.2e-16 ***
Residuals 287.0 45
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
There is a significant difference in words recalled based on process, but why? Investigate with post-hoc tests.
Loading required package: mvtnorm
Loading required package: survival
Loading required package: TH.data
Loading required package: MASS
Attaching package: 'TH.data'
The following object is masked from 'package:MASS':
geyser
<- glht (memory_interactions_young, linfct = mcp (Process = "Tukey" ))summary (comp_young)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = Words ~ Process, data = memory[memory$Age == "Younger",
])
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
Counting - Adjective == 0 -8.300 1.129 -7.349 < 1e-04 ***
Imagery - Adjective == 0 2.800 1.129 2.479 0.11350
Intentional - Adjective == 0 4.500 1.129 3.984 0.00219 **
Rhyming - Adjective == 0 -7.200 1.129 -6.375 < 1e-04 ***
Imagery - Counting == 0 11.100 1.129 9.828 < 1e-04 ***
Intentional - Counting == 0 12.800 1.129 11.333 < 1e-04 ***
Rhyming - Counting == 0 1.100 1.129 0.974 0.86545
Intentional - Imagery == 0 1.700 1.129 1.505 0.56457
Rhyming - Imagery == 0 -10.000 1.129 -8.854 < 1e-04 ***
Rhyming - Intentional == 0 -11.700 1.129 -10.359 < 1e-04 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- single-step method)
Blocking example
Following feather color example from class:
# more than 2? #### <- read.csv ("https://raw.githubusercontent.com/jsgosnell/CUNY-BioStats/master/datasets/wiebe_2002_example.csv" , stringsAsFactors = T)str (feather)
'data.frame': 32 obs. of 3 variables:
$ Bird : Factor w/ 16 levels "A","B","C","D",..: 1 2 3 4 5 6 7 8 9 10 ...
$ Feather : Factor w/ 2 levels "Odd","Typical": 2 2 2 2 2 2 2 2 2 2 ...
$ Color_index: num -0.255 -0.213 -0.19 -0.185 -0.045 -0.025 -0.015 0.003 0.015 0.02 ...
set.seed (25 )<- data.frame (Bird = LETTERS[1 : 16 ], Feather = "Special" , Color_index= feather[feather$ Feather == "Typical" , "Color_index" ] + 3 + runif (16 ,1 ,1 )* .01 )<- merge (feather, special, all = T)Anova (lm (Color_index ~ Feather + Bird, data= feather), type= "III" )
Anova Table (Type III tests)
Response: Color_index
Sum Sq Df F value Pr(>F)
(Intercept) 0.36392 1 59.9538 1.224e-08 ***
Feather 1.67906 2 138.3093 7.208e-16 ***
Bird 0.34649 15 3.8055 0.0008969 ***
Residuals 0.18210 30
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
library (multcomp)<- glht (lm (Color_index ~ Feather + Bird, data= feather), linfct = mcp ("Feather" = "Tukey" ))summary (compare)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = Color_index ~ Feather + Bird, data = feather)
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
Typical - Odd == 0 0.13712 0.02755 4.978 <1e-04 ***
Special - Odd == 0 0.44712 0.02755 16.232 <1e-04 ***
Special - Typical == 0 0.31000 0.02755 11.254 <1e-04 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- single-step method)
#note comparison doesn't work Anova (lm (Color_index ~ Feather * Bird, data= feather), type= "III" )
Error in Anova.lm(lm(Color_index ~ Feather * Bird, data = feather), type = "III"): residual df = 0
Swirl lesson
Swirl is an R package that provides guided lessons to help you learn and review material. These lessons should serve as a bridge between all the code provided in the slides and background reading and the key functions and concepts from each lesson. A full course lesson (all lessons combined) can also be downloaded using the following instructions.
THIS IS ONE OF THE FEW TIMES I RECOMMEND WORKING DIRECTLY IN THE CONSOLE! THERE IS NO NEED TO DEVELOP A SCRIPT FOR THESE INTERACTIVE SESSIONS, THOUGH YOU CAN!
install the “swirl” package
run the following code once on the computer to install a new course
library (swirl)install_course_github ("jsgosnell" , "JSG_swirl_lessons" ) start swirl!
then follow the on-screen prompts to select the JSG_swirl_lessons course and the lessons you want
Here we will focus on the More ANOVAs lesson
TIP: If you are seeing duplicate courses (or odd versions of each), you can clear all courses and then re-download the courses by
exiting swirl using escape key or bye() function
uninstalling and reinstalling courses
uninstall_all_courses ()install_course_github ("jsgosnell" , "JSG_swirl_lessons" ) when you restart swirl with swirl(), you may need to select
No. Let me start something new
Practice
4
Find an example of a factorial ANOVA from a paper that is related to your research or a field of interest. Make sure you understand the connections between the methods, results, and graphs. Briefly answer the following questions
What was the dependent variable?
What were the independent variables?
Was the interaction significant?
If so, how did they interpret findings
If not, were the main effects significant?